Download Demo Data
Download images and tables for the tutorial
After downloading, unzip the folder.
We recommend extracting the CellTune_Store directory inside to your Documents folder.
Important: Avoid extracting to cloud-synced folders (such as Dropbox, OneDrive, or Google Drive), as constant syncing can cause severe slowdowns when generating zarr files during project setup. If you must use a synced folder, disable syncing until the project is fully generated.
Demo Data Contents
The demo dataset included with CellTune provides a complete example of spatial proteomics analysis using multiplexed tissue images from the study: Azulay et al. “A spatial atlas of human gastrointestinal acute GVHD reveals epithelial and immune dynamics underlying disease pathophysiology.” Science Translational Medicine 17.823 (2025).
It consists of MIBI (Multiplexed Ion Beam Imaging) data from duodenum and colon biopsies collected from GVHD patients and matched controls, acquired at 0.4 µm/pixel resolution and exported as 8-bit (0–255) single TIFF images.
Preprocessing followed the MAUI pipeline (Baranski et al., 2021), including background subtraction, noise removal, and aggregate filtering. Segmentation was performed using Mesmer (Greenwald et al., 2022) via the ark-analysis pipeline, using a two-pass strategy designed to better capture goblet cells. For further details, see the methods section of the manuscript.
CellTune_Demo_Data/
Tables/cellTable_features_calculated.parquet
Pre-computed single-cell features, including intensity, morphology, and spatial statistics.CellTypeTable.csv
Metadata describing the cell types.
Labels/Labels.csv
Cell type annotations for landmark cells.
Images/Control_01_FOV_1,Control_01_FOV_2, …,GVHD_03_FOV_2, etc. [Single TIFF folders containing channel images and segmentations.]
The Images/ folder contains 75 subfolders, each corresponding to a single field of view (FOV) from either control or GVHD tissue. Each Image (FOV) folder includes:
-
Protein (Marker) Channels
Single-channel grayscale.tifimages (e.g.,CD3.tif,CD4.tif,CD8.tif,FOXP3.tif, etc.) representing individual protein markers. - Additional Feature Channels
Folders may also include:- Subregion masks (e.g., epithelium, muscle, vasculature)
- Composite or colocalization images (combining multiple marker channels) used for feature extraction
- Segmentation Files
segmentation_labels.tiff: Cell instance segmentation image.
Each pixel is labeled with a unique cell ID, with background as0.segmentation_borders.tiff: Outlines of segmented cell borders (purely for visualization outside of CellTune). These are not required for CellTune (and we usually exclude them in the new project setup).
Each FOV in this dataset has 73 channel files + 2 segmentation files.
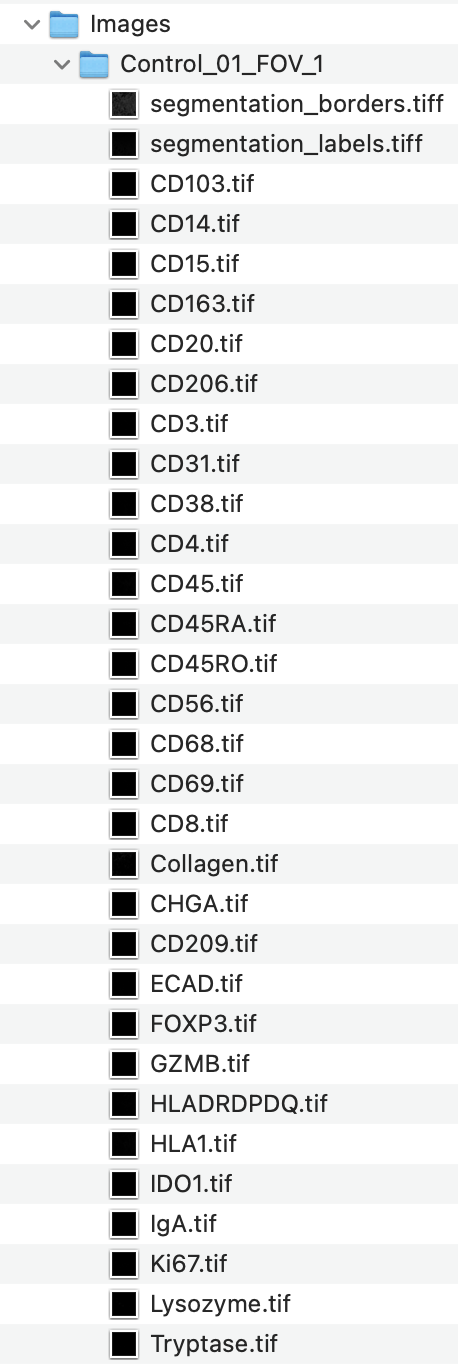
These files will be loaded directly into CellTune when starting a new project.
The following video shows the directory structure and contents of the demo data folder:
Now that you’ve downloaded and extracted the demo data, you’re ready to set up a new project in CellTune.