Marker Table
Preparing the MarkerTable is optional. It usually takes about 10 minutes. We find it helps us to organize our project’s panel and our prior knowledge about expression. It is also great for easily sharing with collaborators. Currently it is for the user’s benefit only, but we plan that CellTune will read and incorporate the table in the future to automate processes - so we highly recommend making it.
Example MarkerTable.csv:
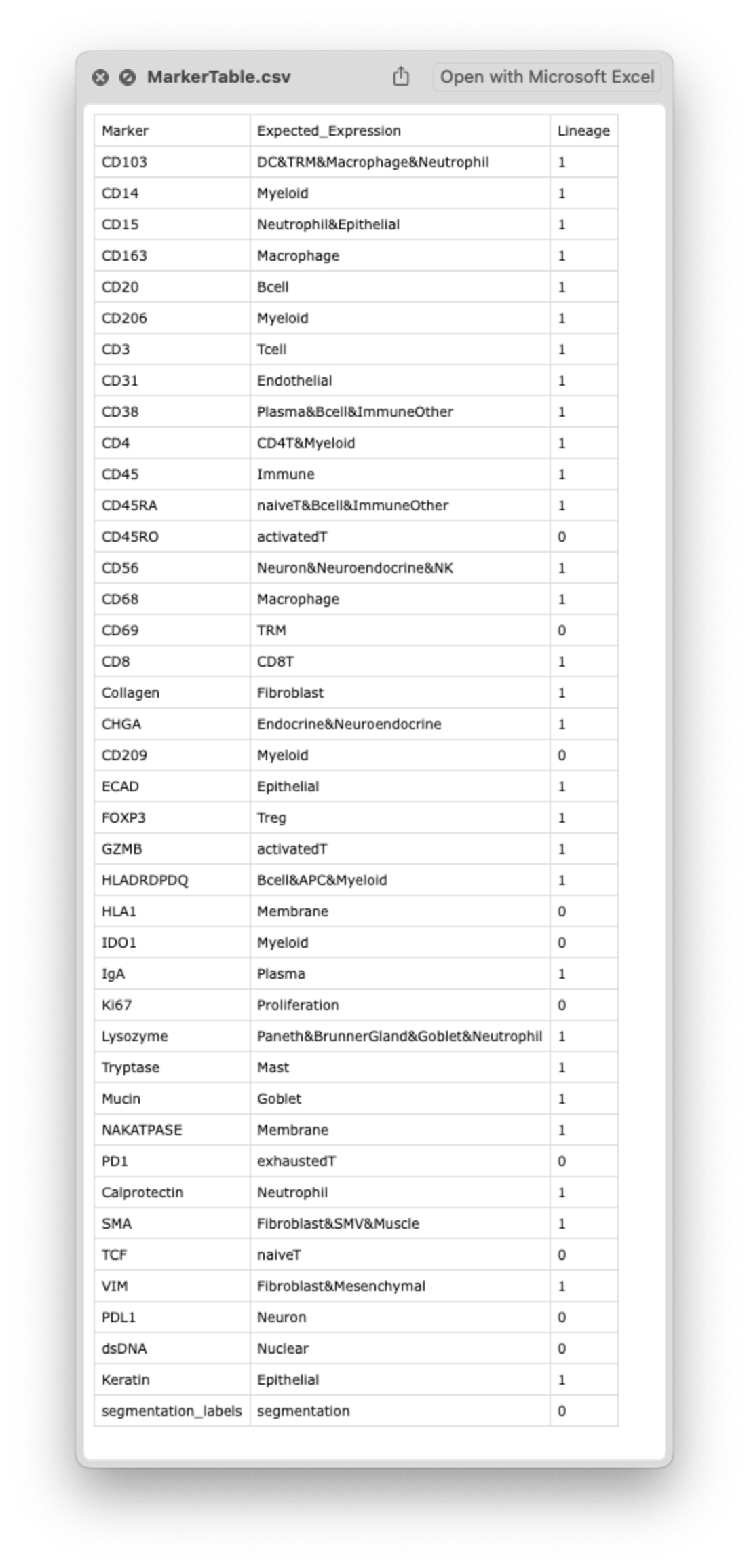
Marker Names
The standardized marker name for each channel. These should be finalized after image organization and follow consistent formatting across datasets. We often include a Marker_orig column to preserve the original vendor or acquisition channel name before applying CellTune naming conventions.
Requirements:
- No spaces, hyphens, or special characters (use underscores sparingly, as needed)
- Consistent naming and capitalization across datasets and batches
Expected Expression
A short list of the cell types and/or functional states where the marker is expected to be expressed. This captures the prior biological knowledge behind including the marker in the panel and is used later when reviewing phenotypes or validating CellTypeTable design.
This information is also very helpful for sharing with collaborators or students who join your project.
Lineage Indicator
Indicates whether this marker is used for cell type (lineage) classification.
1 → Lineage marker
0 → Non-lineage marker
Examples:
CD8 → 1 (used for T-cell lineage classification)
Ki67 → 0 (proliferation marker, not lineage-defining)