Automated Landmarking
Automated Landmarking automatically identifies high-confidence example cells (“landmarks”) for each cell type so you don’t have to manually gate them one by one. It uses the celltype-marker rules table (CellTypeTable) and can operate on the intensity values, marker-positivity probabilities, or both when available. (You must have computed or imported features prior to landmarking.)
The landmark rules are the celltype definitions in the CellTypeTable - it uses the columns PrimaryMarker, SecondaryMarker, and TertiaryMarker. See the documentation for CellTypeTable setup and reference.
Automated Landmarking can be found in the menu under Gating:
You can configure Automated Landmarking using the dialog shown below:
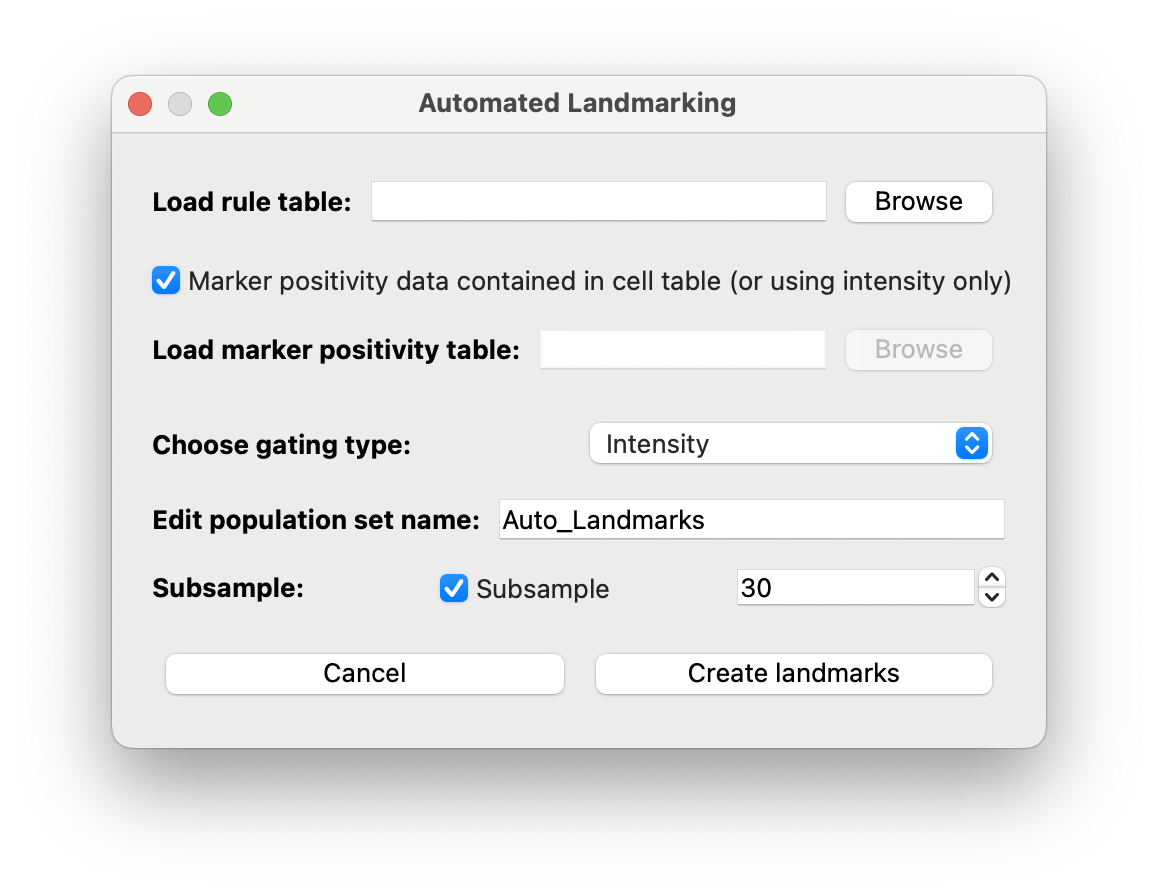
- If you have an external marker positivity table you can upload it from this dialog. The column names in this table must be ‘{MARKER_NAME}__Probability’.
-
Choose gating typeallows you to choose between gating based on:Intensity,Probability,Both. You must upload a marker positivity table to use theProbabilityorBothoptions. - We highly recommend utilizing subsampling, so you can review your landmarks before training. In our manuscript, we showed that there was not any benefit to the final accuracy from initializing with thousands of landmarks.
When the algorithm finishes you will see the following message:
Then you will see a summary of the number of cells landmarked for each celltype.

- You can also see these counts afterwards: From the menu:
Populations -> Population Counts - Remember you need >10 cells per celltype.
What if not enough landmarks are found for a celltype?
Sometimes, the automated landmarking might not find enough or any landmark cells for a celltype. It may be because you did not include some markers in the table as Seconday/Tertiary when they should have been. For example if you did not include CD45 in the CD8T row, then it may not find any CD8T cells since almost all are CD45+. You can adjust the channels and rerun the automated landmarking. Alternatively, you can use the manual landmarking and/or clustering-based landmarking to supplement the automated landmarks. For very rare celltypes with a lot of spillover from neighbors, sometimes it is easier to find some examples doing the manual landmarking just for that one celltype.