Sampling from a Classifier
After training, we can finally sample cells for review & labeling.
- Select your classifier from the classification pane.
-
Click on
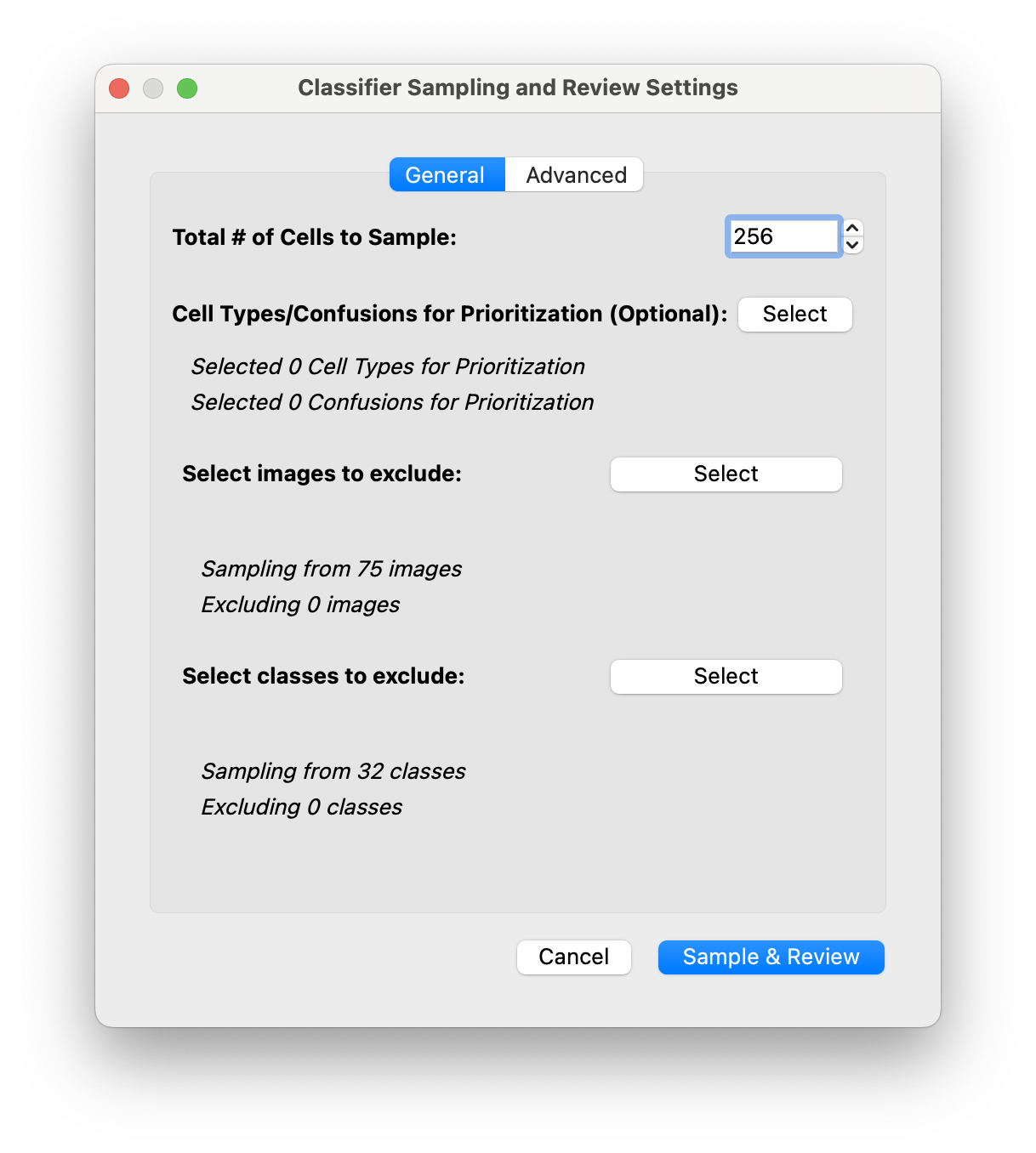
Sample & Reviewin the classification panel.A dialog will open to set the Sampling and Review settings:
-
First you can edit the number of cells to sample. You can sample as many or as few as you’d like, but we found that around 150-300 is optimal to balance how often you train vs label. (Note: You are free to retrain and resample at any point, your labels are always being accumulated!)
-
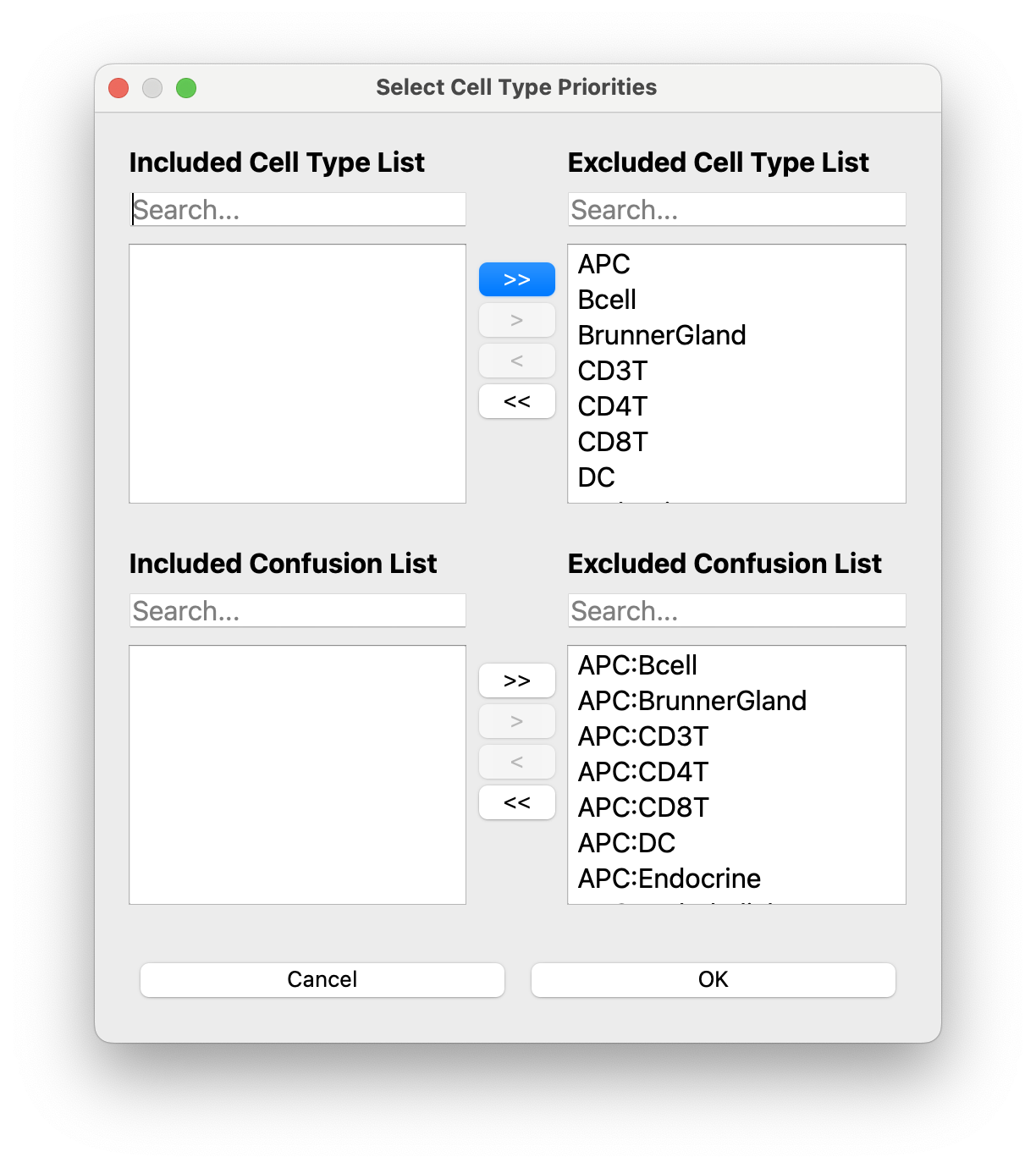
[Optional] You can select specific celltypes or celltype-celltype confusions for prioritization. Note that you will need to change the sampling weighting in the advanced tab!
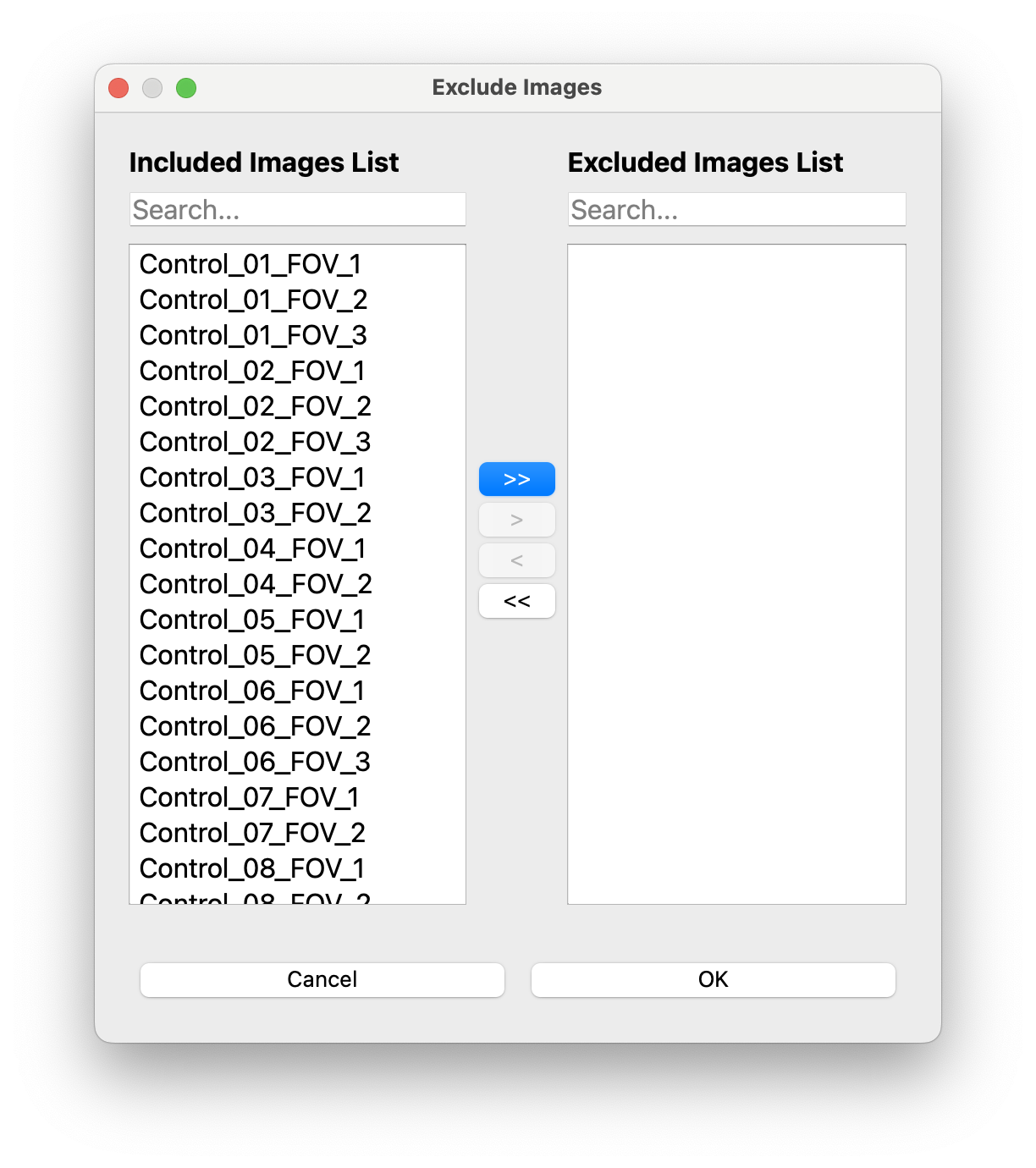
- You can optionally exclude images,
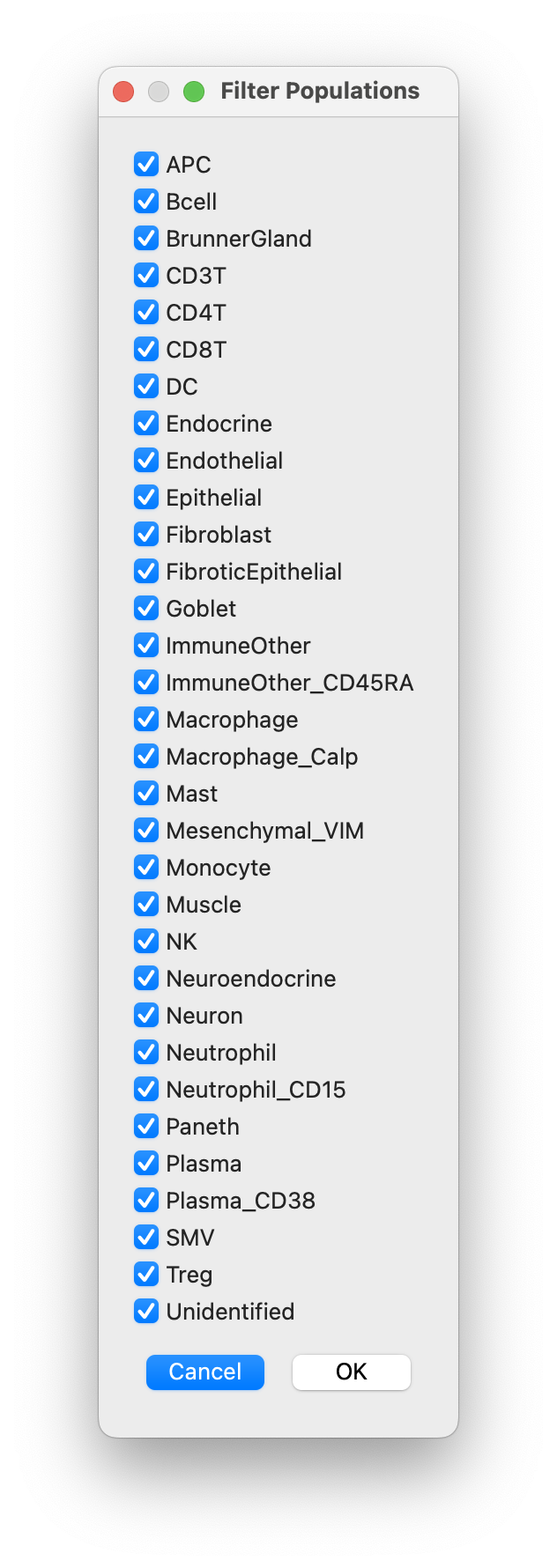
- and/or filter out populations:
- In the advanced tab you can adjust the default sampling weights:
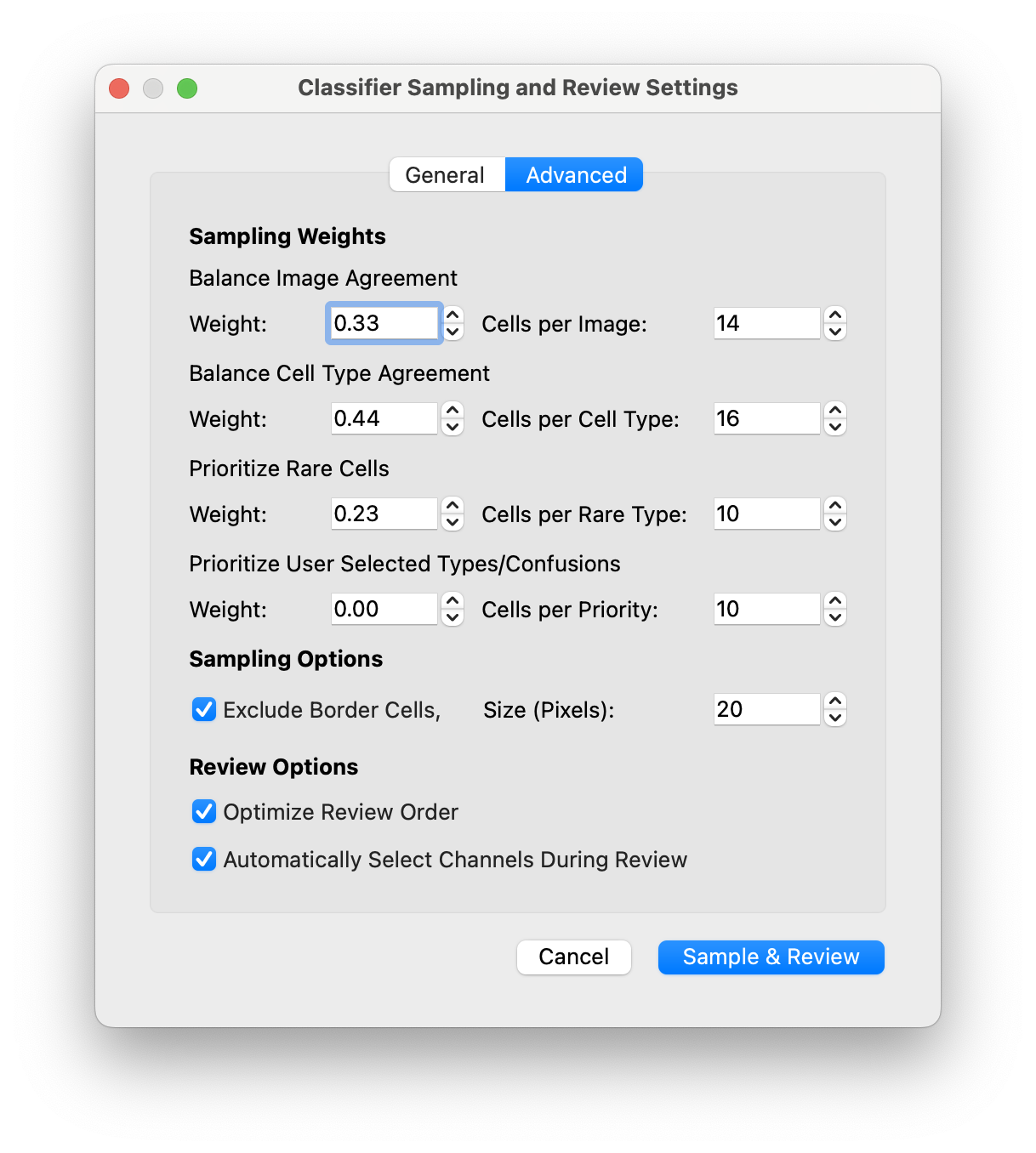
- The weights are the fraction of cells you want to dedicate to each section. For example if you sample 256 cells and set the weight for Balancing Cell Type Agreement = 0.5, then 128 cells will be sampled for this task.
- The numbers on the right indicate how many cells to sample for each of the top most disagreeing cell types
i.e. CellTune ranks the celltypes, then samples 16 cells from each cell type starting from the one with the lowest agreement until it reaches 128 cells total.
- Optionally you can exclude cells on the borders of the images (often we get some artifact staining on the edges and don’t want to label these cells)
-
Review Settings:
- Optimize Review Order = orders the sampled cells by their confusions so you get all the Bcell-CD4T cell confusions one after the other, to minimize the amount of context switching you have to do as you navigate from one cell to the next.
- Automatically Select Channels During Review = Uses the population info (stored and imported from the CellTypeTable) to put on the top 3 channels associated with the predicted celltypes. Remaining channels are filled by the highest expressing proteins on the cell. (In some versions we reserved one channel for the nuclear marker, DAPI). You can turn this setting off at any time during the review.