CellType Table
The CellTypeTable can be used for automated landmarking and automated channel selection during cell review.*
Example CellTypeTable.csv:
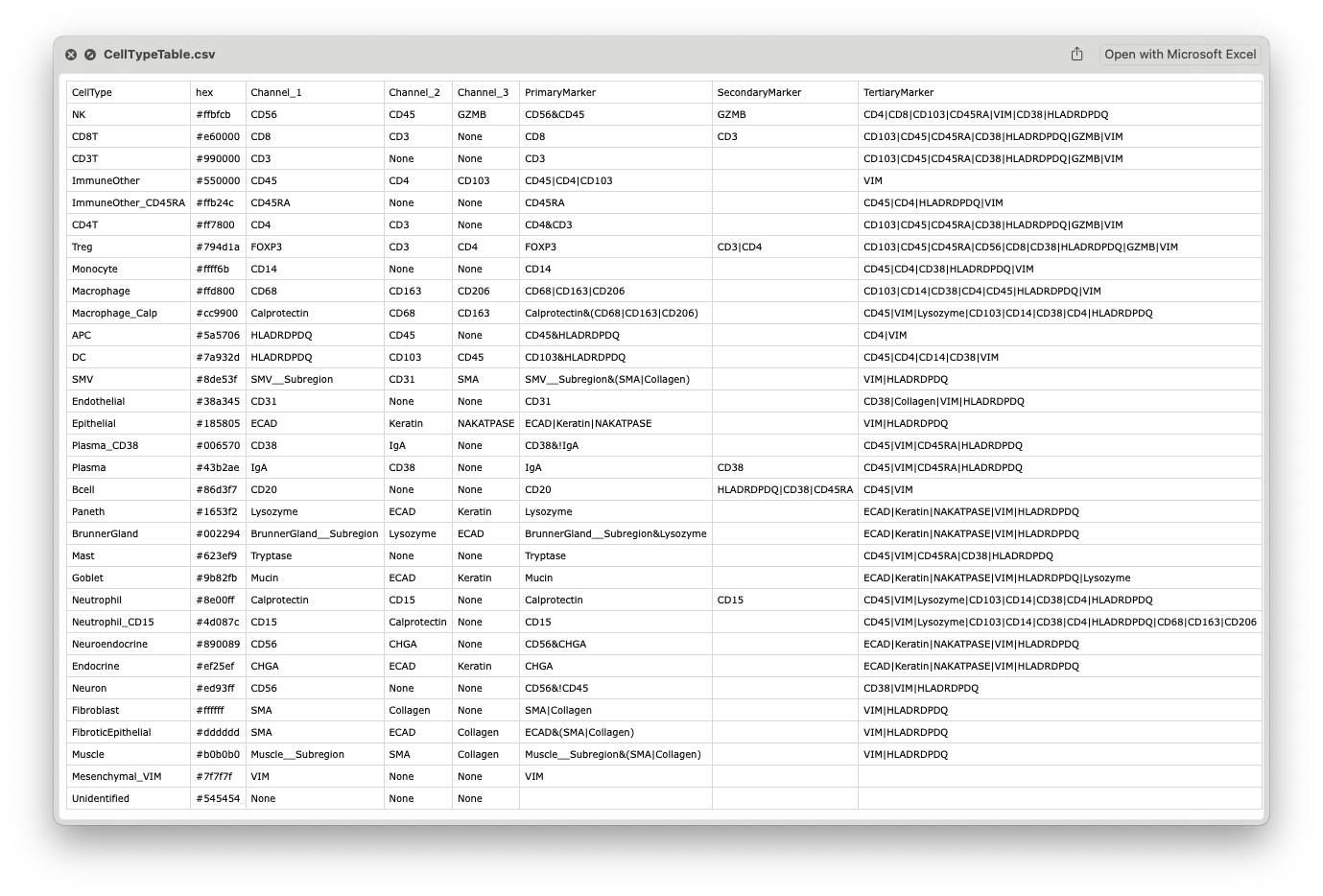
Example table can be downloaded from the demo data of the tutorials.
Suggested Location: CellTune_Store/CellTune_Data/{PROJ_NAME}/Tables/
Cell Type Names
Column name: CellType (or class)
Description: The assigned cell type or lineage name for each cell. CellTune users are required to follow a strict naming convention (Just kidding, you can input whatever you want, but we do make everyone in our lab follow these guidelines:
- Capitalize names
- Only letters and underscores
- Avoid spaces and special characters (+, -, parentheses, punctuation)
- Avoid pluralizing celltype names (Neutrophil, not Neutrophils)
- Avoid using the word
cells(except for 1 letter celltypes, e.g. B & T cells are Bcell and Tcell) - Use celltype name first before any subset protein identifiers (e.g. Macrophage_Calp instead of Calp_Macrophage for Calprotectin+ Macrophages)
When defining subsets, we strongly prefer naming by positive markers whenever possible. For example: Plasma_CD38 = Plasma cells that are CD38⁺ and IgA⁻ Plasma or Plasma_IgA = IgA⁺ Plasma cells If possible avoid negative-marker naming like IgA_neg_Plasma, Plasma_IgA-, Plasma_IgA_neg, etc. Using positive markers keeps labels intuitive—when you see Plasma_CD38, you immediately know you’re looking for CD38⁺ cells.
See our CellTuneDepot for a curated database of recommended celltype names for classification in CellTune.
On a serious note: Compact, informative names make classification and review much easier. For example, use CD8T, not “CD8+ T cells”—trust us, this will make your life much happier. When writing and preparing figures for publication, you will change all the names to your preferred presentation style anyway (e.g., adding superscripts).
Above all, be consistent with your naming in CellTune. We don’t tolerate anything less (…just kidding…but not really).
Primary Marker Column
Column name: PrimaryMarker
Description: The minimal expression that defines membership to the celltype. These must be expressed. For example, in most datasets CD8+CD3- cells are classified as CD8T, so CD8 is the PrimaryMarker. Whereas CD4+CD3- could be myeloid cells, thus CD3&CD4 is the PrimaryMarker expression for CD4T.
- You can use AND (
&), OR (|), and NOT (!) operators as well as parentheses. - Do not use spaces.
- Not is assumed for all channels not present in Primary, Secondary, and Tertiary columns, but is often helpful to include in the definition for user clarity. E.g.
CD38&!IgAfor Plasma_CD38 (CD38+IgA- Plasma cells), andIgAfor Plasma)
Example using OR operator: Calprotectin&(CD163|CD68|CD206) = Macrophage_Calp.
Additional examples can be seen in the example table above.
Secondary & Tertiary Marker Columns
Column names: SecondaryMarker and TertiaryMarker
Description: Markers that could be expressed but are not required for membership to the celltype. E.g. CD8+CD45- cells will be classified as CD8T as will CD8+CD45+ cells. So they are not required but present (and usually present).
‘SecondaryMarker’ designation is given to TertiaryMarkers that are very specific for a celltype, it is purely for organization (the user). E.g. CD3 is very specific for CD8 and we expect it on all/most CD8T. So we include it there. A similar argument can be made for CD45, but we prefer to put it as Tertiary for all the immune cell types (aside from those where it is Primary). See the example table above.
Some proteins stain the tissue broadly and all cells have some signal of expression from them. For example Collagen often falls into this category. In this case we include this marker as a TertiaryMarker as if we set strict gates against it we might not find any of the desired cells.
Associated Channel Columns
Column names: Channel_1, Channel_2, Channel_3
Description: These are the top 3 channels associated with the celltype, the channels you would want to put on when evaluating/reviewing. These channels will be used for automated channel selection in Review.
Color Column
Column name: hex
Description:
Cell Type Colors!
As you can see, we feel very strongly about celltype colors. We spent a lot of time generating a color palette of over 30 colors we can differentiate easily between. Then we made a default assignment for the major cell types.
CellTune CytoPalette
We use this as a starting point for our projects, and then make minimal adjustments to it - adding the cell types that aren’t present and swapping some assignments if there is something specific we want to contrast even more. e.g. if our project was specifically about Neutrophils in the Tumor we might want to make them further apart, instead of Blue and Purple we might move the Neutrophils to be Pink or Orange.